在MATLAB中更改seqlogo数字的x轴
我正在以编程方式制作大量seqlogos。它们有数百列宽,因此运行seqlogo通常会创建太薄而无法看到的字母。我注意到我只关心这些列中的一些(不一定是连续的列)......大多数是噪音但有些是高度保守的。
我使用的代码如下:
wide_seqs = cell2mat(arrayfun(@randseq, repmat(200, [500 1]), 'uniformoutput', false));
wide_seqs(:, [17,30, 55,70,130]) = repmat(['ATCGG'], [500 1])
conserve_cell = seqlogo(wide_seqs, 'displaylogo', false);
high_bit_cols = any(conserve_cell{2}>1.0,1);
[~, handle] = seqlogo(wide_seqs(:,high_bit_cols ));
虽然当我这样做时,我会丢失有关数据来自哪些列的信息。
通常我只会改变seqlogo的x轴。但是,seqlogo是某种疯狂的基于java的对象,并且调用如下:
set(handle, 'xticklabel', num2str(find(high_bit_cols)))
不行。任何帮助将不胜感激。
谢谢, 将
编辑:
在赏金上我愿意接受任何一种改变轴标签的疯狂方法包括(但不限于):使用图像处理工具箱在保存后修改图像,使用文本框创建新的seqlogo函数,修改java代码(如果可能的话),等等。我不愿意接受“使用python”,“使用这个R库”或任何其他类型的非Matlab解决方案。
5 个答案:
答案 0 :(得分:5)
好的,我这个问题杀了几个小时。您似乎无法在该hgjavacomponent对象的顶部放置任何MATLAB对象(轴或文本框)。当然,我无法修改java代码。所以我找到的唯一可行解决方案是从头开始创建数字。
我不想重写代码来计算权重矩阵(符号高度),你已经这样做了。但是如果你根本不想使用MATLAB的seqlogo,那就可以了。所以我已经改变了你的最后一行以获得矩阵:
[wm, handle] = seqlogo(wide_seqs(:,high_bit_cols ));
文本符号的问题在于您无法准确控制其大小,无法使符号适合文本框。这可能就是MATLAB决定使用java图形对象的原因。但我们可以创建符号图像并处理它们。
以下是创建字母图像的代码:
letters = wm{1};
clr = [0 1 0; 0 0 1; 1 0.8 0;1 0 0]; % corresponding colors
for t = 1:numel(letters)
hf = figure('position',[200 200 100 110],'color','w');
ha = axes('parent',hf, 'visible','off','position',[0 0 1 1]);
ht = text(50,55,letters(t),'color',clr(t,:),'units','pixels',...
'fontsize',100,'fontweight','norm',...
'vertical','mid','horizontal','center');
F = getframe(hf); % rasterize the letter
img = F.cdata;
m = any(img < 255,3); % convert to binary image
m(any(m,2),any(m,1))=1; % mask to cut white borders
imwrite(reshape(img(repmat(m,[1 1 3])),[sum(any(m,2)) sum(any(m,1)) 3]),...
[letters(t) '.png'])
close(hf)
end
然后我们使用这些图像绘制新的seqlogo图:
xlabels = cellstr(num2str(find(high_bit_cols)'));
letters = wm{1};
wmat=wm{2}; % weight matrix from seqlogo
[nletters npos] = size(wmat);
wmat(wmat<0) = 0; % cut negative values
% prepare the figure
clf
hAx = axes('parent',gcf,'visible','on');
set(hAx,'XLim',[0.5 npos+0.5],'XTick',1:npos,'XTickLabel',xlabels)
ymax = ceil(max(sum(wmat)));
ylim([0 ymax])
axpos = get(hAx,'Position');
step = axpos(3)/npos;
% place images of letters
for i=1:npos
[wms idx] = sort(wmat(:,i)); % largest on the top
let_show = letters(idx);
ybot = axpos(2);
for s=1:nletters
if wms(s)==0, continue, end;
axes('position',[axpos(1) ybot step wms(s)/ymax*axpos(4)])
ybot = ybot + wms(s)/ymax*axpos(4);
img = imread([let_show(s) '.png']);
image(img)
set(gca,'visible','off')
end
axpos(1)=axpos(1)+step;
end
结果如下: alt text http://img716.imageshack.us/img716/2073/seqlogoexample.png
当然,代码和图形可以进一步改进,但我希望这是你可以开始使用的东西。如果我错过了什么,请告诉我。
答案 1 :(得分:4)
我遇到了同样的问题yuk did试图修改SEQLOGO中的数字,所以这是我尝试自己的版本来模仿它的外观。这是一个函数seqlogo_new.m,你给出两个参数:你的序列和一个可选的最小位值。它需要一个可以找到at this link的图像文件ACGT.jpg。
以下是该功能的代码:
function hFigure = seqlogo_new(S,minBits)
%# SEQLOGO_NEW Displays sequence logos for DNA.
%# HFIGURE = SEQLOGO_NEW(SEQS,MINBITS) displays the
%# sequence logo for a set of aligned sequences SEQS,
%# showing only those columns containing at least one
%# nucleotide with a minimum bit value MINBITS. The
%# MINBITS parameter is optional. SEQLOGO_NEW returns
%# a handle to the rendered figure HFIGURE.
%#
%# SEQLOGO_NEW calls SEQLOGO to perform some of the
%# computations, so to use this function you will need
%# access to the Bioinformatics Toolbox.
%#
%# See also seqlogo.
%# Author: Ken Eaton
%# Version: MATLAB R2009a
%# Last modified: 3/30/10
%#---------------------------------------------------------
%# Get the weight matrix from SEQLOGO:
W = seqlogo(S,'DisplayLogo',false);
bitValues = W{2};
%# Select columns with a minimum bit value:
if nargin > 1
highBitCols = any(bitValues > minBits,1); %# Plot only high-bit columns
bitValues = bitValues(:,highBitCols);
else
highBitCols = true(1,size(bitValues,2)); %# Plot all columns
end
%# Sort the bit value data:
[bitValues,charIndex] = sort(bitValues,'descend'); %# Sort the columns
nSequence = size(bitValues,2); %# Number of sequences
maxBits = ceil(max(bitValues(:))); %# Upper plot limit
%# Break 4-letter image into a 1-by-4 cell array of images:
imgACGT = imread('ACGT.jpg'); %# Image of 4 letters
[nRows,nCols,nPages] = size(imgACGT); %# Raw image size
letterIndex = round(linspace(1,nCols+1,5)); %# Indices of letter tile edges
letterImages = {imgACGT(:,letterIndex(1):letterIndex(2)-1,:), ...
imgACGT(:,letterIndex(2):letterIndex(3)-1,:), ...
imgACGT(:,letterIndex(3):letterIndex(4)-1,:), ...
imgACGT(:,letterIndex(4):letterIndex(5)-1,:)};
%# Create the image texture map:
blankImage = repmat(uint8(255),[nRows round(nCols/4) 3]); %# White image
fullImage = repmat({blankImage},4,2*nSequence-1); %# Cell array of images
fullImage(:,1:2:end) = letterImages(charIndex); %# Add letter images
fullImage = cat(1,cat(2,fullImage{1,:}),... %# Collapse cell array
cat(2,fullImage{2,:}),... %# to one 3-D image
cat(2,fullImage{3,:}),...
cat(2,fullImage{4,:}));
%# Initialize coordinates for the texture-mapped surface:
X = [(1:nSequence)-0.375; (1:nSequence)+0.375];
X = repmat(X(:)',5,1); %'# Surface x coordinates
Y = [zeros(1,nSequence); cumsum(flipud(bitValues))];
Y = kron(flipud(Y),[1 1]); %# Surface y coordinates
Z = zeros(5,2*nSequence); %# Surface z coordinates
%# Render the figure:
figureSize = [602 402]; %# Figure size
screenSize = get(0,'ScreenSize'); %# Screen size
offset = (screenSize(3:4)-figureSize)/2; %# Offset to center figure
hFigure = figure('Units','pixels',...
'Position',[offset figureSize],...
'Color',[1 1 1],...
'Name','Sequence Logo',...
'NumberTitle','off');
axes('Parent',hFigure,...
'Units','pixels',...
'Position',[60 100 450 245],...
'FontWeight','bold',...
'LineWidth',3,...
'TickDir','out',...
'XLim',[0.5 nSequence+0.5],...
'XTick',1:nSequence,...
'XTickLabel',num2str(find(highBitCols)'),... %'
'YLim',[-0.03 maxBits],...
'YTick',0:maxBits);
xlabel('Sequence Position');
ylabel('Bits');
surface(X,Y,Z,fullImage,...
'FaceColor','texturemap',...
'EdgeColor','none');
view(2);
end
以下是一些使用示例:
S = ['ATTATAGCAAACTA'; ... %# Sample data
'AACATGCCAAAGTA'; ...
'ATCATGCAAAAGGA'];
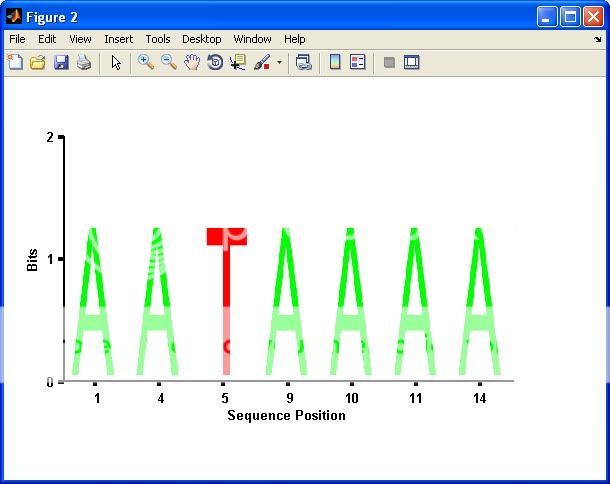
seqlogo_new(S); %# A normal plot similar to SEQLOGO
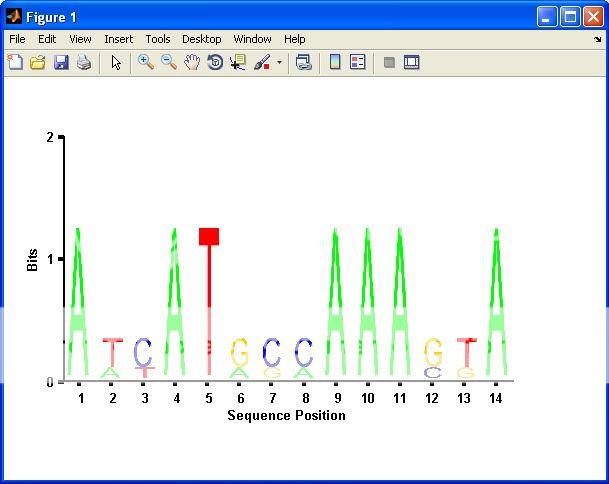
seqlogo_new(S,1); %# Plot only columns with bits > 1
答案 2 :(得分:3)
所以我用yuk和gnovice的解决方案创建了另一个解决方案。当我玩解决方案时,我意识到我真的希望能够将输出用作“子图”并能够任意改变字母的颜色。
由于yuk使用了嵌入字母的programitically放置轴对象,修改他的代码以绘制成任意轴对象将是非常烦人的(尽管不是不可能)。由于gnovice的解决方案从预先创建的文件中读取字母,因此很难修改代码以针对任意颜色方案或字母选择运行。所以我的解决方案使用yuk解决方案中的“字母生成”代码和gnovice解决方案中的“图像叠加”方法。
还有大量的参数解析和检查。以下是我的综合解决方案......我只是为了完整性而将其包括在内,我显然无法赢得自己的赏金。我会让社区决定奖励,并在时间限制结束时给予最高评级的奖励......如果出现平局,我会把它交给代表最低的人(他们可能“需要”更多)。
function [npos, handle] = SeqLogoFig(SEQ, varargin)
% SeqLogoFig
% A function which wraps around the bioinformatics SeqLogo command
% and creates a figure which is actually a MATLAB figure. All
% agruements for SEQLOGO are passed along to the seqlogo calculation.
% It also supports extra arguements for plotting.
%
% [npos, handle] = SeqLogoFig(SEQ);
%
% SEQ A multialigned set of sequences that is acceptable
% to SEQLOGO.
% npos The positions that were actually plotted
% handle An axis handle to the object that was plotted.
%
% Extra Arguements:
%
% 'CUTOFF' A bit-cutoff to use for deciding which columns to
% plot. Any columns that have a MAX value which is
% greater than CUTOFF will be provided. Defaults to
% 1.25 for NT and 2.25 for AA.
%
% 'TOP-N' Plots only the top N columns as ranked by thier MAX
% bit conservation.
%
% 'AXES_HANDLE' An axis handle to plot the seqlogo into.
%
% 'INDS' A set of indices to to plot. This overrides any
% CUTOFF or TOP-N that were provided
%
%
%
%
%% Parse the input arguements
ALPHA = 'nt';
MAX_BITS = 2.5;
RES = [200 80];
CUTOFF = [];
TOPN = [];
rm_inds = [];
colors = [];
handle = [];
npos = [];
for i = 1:2:length(varargin)
if strcmpi(varargin{i}, 'alphabet')
ALPHA = varargin{i+1};
elseif strcmpi(varargin{i}, 'cutoff')
CUTOFF = varargin{i+1};
%we need to remove these so seqlogo doesn't get confused
rm_inds = [rm_inds i, i+1]; %#ok<*AGROW>
elseif strcmpi(varargin{i}, 'colors')
colors = varargin{i+1};
rm_inds = [rm_inds i, i+1];
elseif strcmpi(varargin{i}, 'axes_handle')
handle = varargin{i+1};
rm_inds = [rm_inds i, i+1];
elseif strcmpi(varargin{i}, 'top-n')
TOPN = varargin{i+1};
rm_inds = [rm_inds i, i+1];
elseif strcmpi(varargin{i}, 'inds')
npos = varargin{i+1};
rm_inds = [rm_inds i, i+1];
end
end
if ~isempty(rm_inds)
varargin(rm_inds) = [];
end
if isempty(colors)
colors = GetColors(ALPHA);
end
if strcmpi(ALPHA, 'nt')
MAX_BITS = 2.5;
elseif strcmpi(ALPHA, 'aa')
MAX_BITS = 4.5;
end
if isempty(CUTOFF)
CUTOFF = 0.5*MAX_BITS;
end
%% Calculate the actual seqlogo.
wm = seqlogo(SEQ, varargin{:}, 'displaylogo', false);
%% Generate the letters
letters = wm{1};
letter_wins = cell(size(letters));
[~, loc] = ismember(letters, colors(:,1));
loc(loc == 0) = size(colors,1);
clr = cell2mat(colors(loc, 2)); % corresponding colors
for t = 1:numel(letters)
hf = figure('position',[200 200 100 110],'color','w');
ha = axes('parent',hf, 'visible','off','position',[0 0 1 1]);
ht = text(50,55,letters(t),'color',clr(t,:),'units','pixels',...
'fontsize',100,'fontweight','norm',...
'vertical','mid','horizontal','center');
F = getframe(hf); % rasterize the letter
img = F.cdata;
m = any(img < 255,3); % convert to binary image
m(any(m,2),any(m,1))=1; % mask to cut white borders
letter_wins{t} = reshape(img(repmat(m,[1 1 3])),[sum(any(m,2)) sum(any(m,1)) 3]);
close(hf);
end
%% Use the letters to generate a figure
%create a "image" that will hold the final data
wmat = wm{2};
if isempty(npos)
if isempty(TOPN)
npos = find(any(wmat>CUTOFF,1));
else
[~, i] = sort(max(wmat,[],1), 'descend');
npos = sort(i(1:TOPN));
end
end
fig_data = 255*ones(RES(1), RES(2)*(length(npos)+1)+length(npos)*2,3);
bitscores = linspace(0, MAX_BITS, size(fig_data,1));
tick_pos = zeros(length(npos),1);
% place images of letters
for i=1:length(npos)
[wms idx] = sort(wmat(:,npos(i)), 'descend'); % largest on the top
bits = [flipud(cumsum(flipud(wms))); 0];
let_data = letter_wins(idx(wms>0));
for s=1:length(let_data)
start_pos = find(bitscores>=bits(s),1);
end_pos = find(bitscores<=bits(s+1),1, 'last');
if isempty(start_pos) || isempty(end_pos) || end_pos > start_pos
continue
end
img_win = imresize(let_data{s}, [start_pos-end_pos, RES(2)]);
fig_data(start_pos-1:-1:end_pos, (i*RES(2)-RES(2)*.5:i*RES(2)+RES(2)*.5-1)+2*i,:) = img_win;
end
tick_pos(i) = i*RES(2)+2*i;
end
if ~isempty(handle)
image(handle,[0 size(fig_data,2)], [0 MAX_BITS],fig_data./255)
else
handle = image([0 size(fig_data,2)], [0 MAX_BITS],fig_data./255);
end
set(gca, 'ydir', 'normal', 'xtick', tick_pos, ...
'userdata', tick_pos, 'xticklabel', npos);
xlabel('position')
ylabel('bits')
function colors = GetColors(alpha)
% get the standard colors for the sequence logo
if strcmpi(alpha, 'nt')
colors = cell(6,2);
colors(1,:) = {'A', [0 1 0]};
colors(2,:) = {'C', [0 0 1]};
colors(3,:) = {'G', [1 1 0]};
colors(4,:) = {'T', [1 0 0]};
colors(5,:) = {'U', [1 0 0]};
colors(6,:) = {'', [1 0 1]};
elseif strcmpi(alpha, 'aa')
colors = cell(21,2);
colors(1,:) = {'G', [0 1 0]};
colors(2,:) = {'S', [0 1 0]};
colors(3,:) = {'T', [0 1 0]};
colors(4,:) = {'Y', [0 1 0]};
colors(5,:) = {'C', [0 1 0]};
colors(6,:) = {'Q', [0 1 0]};
colors(7,:) = {'N', [0 1 0]};
colors(8,:) = {'A', [1 165/255 0]};
colors(9,:) = {'V', [1 165/255 0]};
colors(10,:) = {'L', [1 165/255 0]};
colors(11,:) = {'I', [1 165/255 0]};
colors(12,:) = {'P', [1 165/255 0]};
colors(13,:) = {'W', [1 165/255 0]};
colors(14,:) = {'F', [1 165/255 0]};
colors(15,:) = {'M', [1 165/255 0]};
colors(16,:) = {'D', [1 0 0]};
colors(17,:) = {'E', [1 0 0]};
colors(18,:) = {'K', [0 0 1]};
colors(19,:) = {'R', [0 0 1]};
colors(20,:) = {'H', [0 0 1]};
colors(21,:) = {'', [210/255 180/255 140/255]};
else
error('SeqLogoFigure:BADALPHA', ...
'An unknown alphabet was provided: %s', alpha)
end
我已经将它提交给了Mathworks FileExchange ...当它被批准后我会发布一个链接。
我唯一感到烦恼的是,因为它创建了字母图像,它可以快速显示小图窗口。如果有人知道一个可以避免的技巧,我很乐意听到它。
编辑:Mathworks已批准我提交的文件...您可以在FileExchange下载它:http://www.mathworks.com/matlabcentral/fileexchange/27124
答案 3 :(得分:1)
关于x轴,似乎图中没有标准轴(findobj(handle,'type','axes')为空),而是类 com.mathworks.toolbox.bioinfo.sequence.SequenceLogo <的自定义对象 ...
在不相关的说明中,您可以通过更简单的调用替换您的第一行:
wide_seqs = reshape(randseq(200*500),[],200);
答案 4 :(得分:0)
如果轴是java对象,那么您可能希望使用uiinspect查看其方法和属性。这可能会让你知道你应该编辑什么来获得你想要的行为(不幸的是,我没有工具箱,所以我无法为你查找)。
- 我写了这段代码,但我无法理解我的错误
- 我无法从一个代码实例的列表中删除 None 值,但我可以在另一个实例中。为什么它适用于一个细分市场而不适用于另一个细分市场?
- 是否有可能使 loadstring 不可能等于打印?卢阿
- java中的random.expovariate()
- Appscript 通过会议在 Google 日历中发送电子邮件和创建活动
- 为什么我的 Onclick 箭头功能在 React 中不起作用?
- 在此代码中是否有使用“this”的替代方法?
- 在 SQL Server 和 PostgreSQL 上查询,我如何从第一个表获得第二个表的可视化
- 每千个数字得到
- 更新了城市边界 KML 文件的来源?